Publications
2025
Romero-Pérez, P. S.; Moran, H. M.; Horani, A.; Truong, A.; Manriquez-Sandoval, E.; Ramirez, J. F.; Martinez, A.; Gollub, E.; Hunter, K.; Lotthammer, J. M.; Emenecker, R.; Liu, H.; Iwasa, J. H.; Boothby, T. C.; Holehouse, A. S.; Fried, S. D.*; Sukenik, S.
“Protein Surface Chemistry Encodes an Adaptive Tolerance to Desiccation”. bioRxiv 2024. DOI: 10.1101/2024.07.28.604841v2. Cell Systems 2025. Accepted.
Vu, Q. V.; Sitarik, I.; Jiang, Y.; Xia, Y.; Sharma, P.; Yadav, D.; Song, H.; Li, M. S.; Fried, S. D.; O’Brien, E. P.
“Non-native entanglement protein misfolding observed in all-atom simulations and supported by experimental structural ensembles” Science Advances 2025, 11, 33, adt8974.
Makarov, M.; Kryštůfek, R.; Manriquez-Sandoval, E.; Dutta, S.; Kalvoda, T.; Kormanik, J. M.; Chervanets, T.; Lébl, M.; Brown, S.; Singharoy, A.; Fried, S. D.; Hlouchova, K
“A Primordial Acidic Amino Acid Alphabet Plausibly Served as a Stepping Stone to Foldable Polymers” Protein Science 2025. Under Review.
Jiang, Y.; Xia, Y.; Sitarik, I.; Sharma, P.; Song, H.; Fried, S. D.; O’Brien, E. P.
“Protein misfolding involving entanglements provides a structural explanation for the origin of stretched-exponential refolding kinetics” Science Advances 2025, 11, 11, ads7379.
Liu, H.; Cai, X.; Shi, L.; Pillai, M.; Das, N.; Tarbox, H. E.; Ge, Y.; Yue, K.; Yang, X.; Rath, P.; Badiee, M.; Fabilane, C. S.; Spangler, J. B.; Bedford, M. T.; Myong, S.; Fried, S. D.; Ding, X.; Leung, A. K. L.
“Transient Poly(ADP-Ribose) Triggers FUS Condensation Hysteresis via a Prion-Like Mechanism” Molecular Cell 2025. Under Review.
Faustino, A. M.; Nune, M.; Merino-Urteaga, R.; Manriquez-Sandoval, E.; Poyton, M.; Ha, T.; Wolberger, C.; Fried, S. D.*
“An Intrinsically Disordered Region of Ubp10 Regulates its Binding and Activity on Ubiquitinated Histone Substrates” Protein Science 2025, 34, 8, e70237.
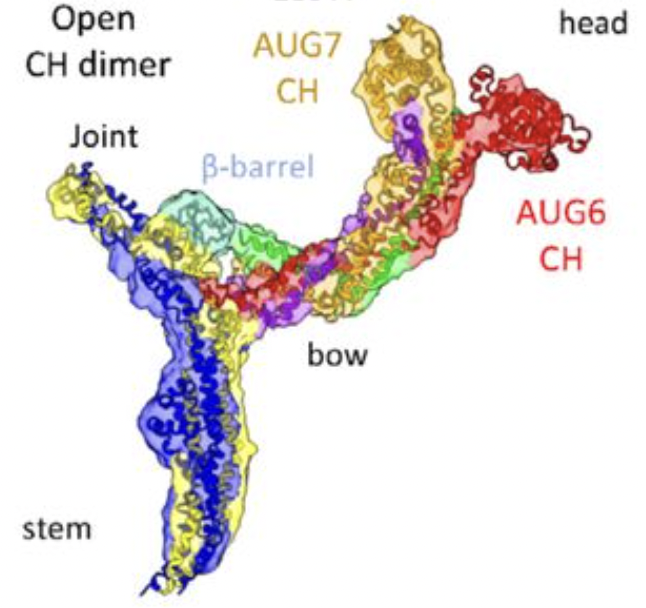
Ashaduzzaman, M.; Taheri, A.; Lee, Y. R.; Tang, Y.; Guo, F.; Fried, S. D.; Liu, B.; Al-Bassam, J.
“Cryo-EM structures of the Plant Augmin reveal its intertwined coiled-coil assembly, antiparallel dimerization and NEDD1 binding mechanisms.” bioRxiv 2025. DOI: 10.1101/2025.02.25.640204. Nature Comm. 2025. In Revision.
Rappsilber, J.; Bruce, J.; Combe, C.; Fried, S. D.; Heck, A. J. R.; Iacobucci, C.; Leitner, A; Mechtler, K.; Novak, P.; O'Reilly, F.; Schriemer, D. C.; Sinz, A.; Stengel, F.; Thalassino, K.
“A Roadmap for Improving Data Sharing in Crosslinking Mass Spectrometry” Mol. Cell. Proteomics 2025, 24, 8, 101024.
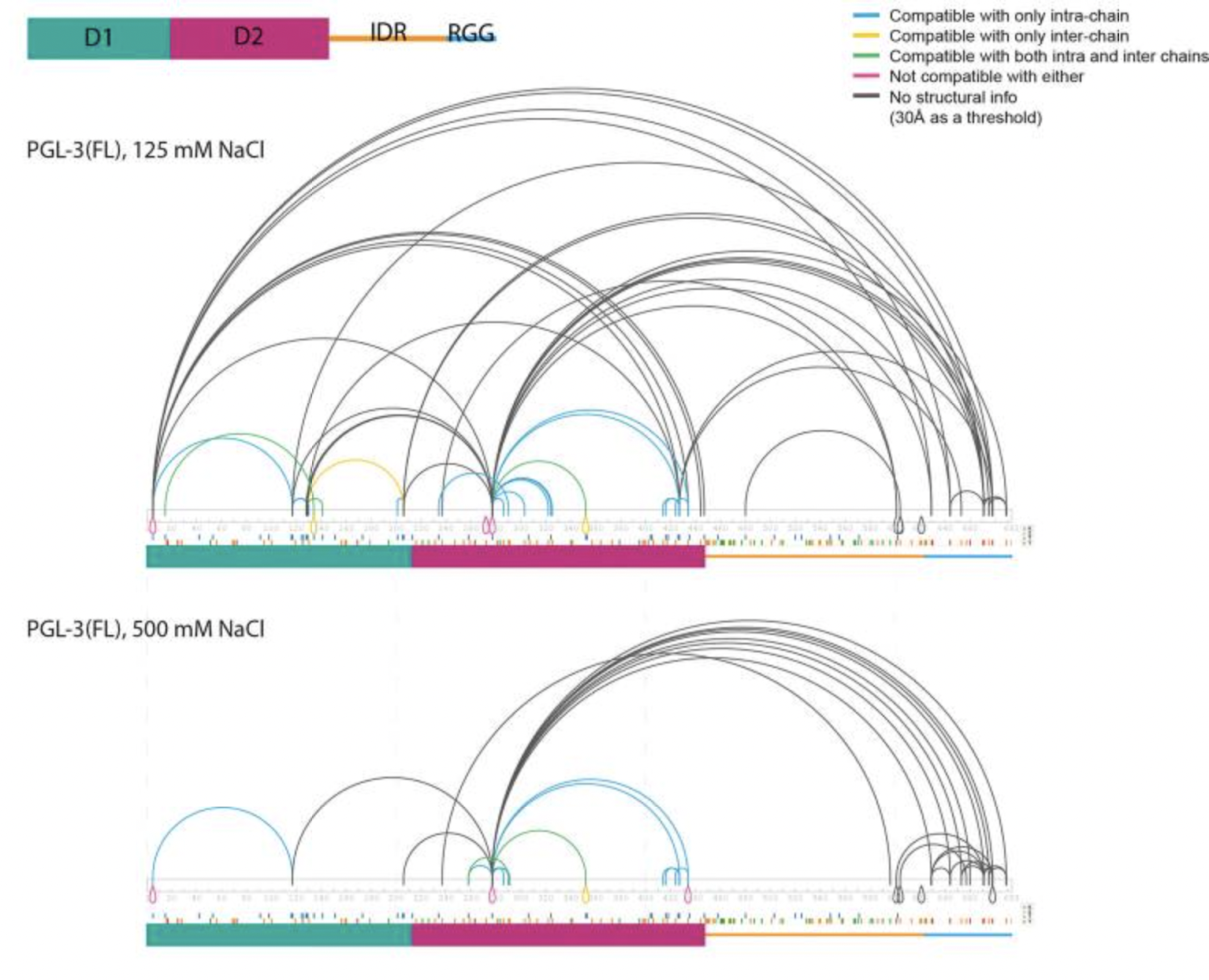
Kuroiwa, R.; Sharma, P.; Putnam, A.; Fried, S. D.; Seydoux, G.
“Phase separation of PGL-3 is driven by oligomerization of ordered domains and modulated by heterotypic interactions between ordered and disordered domains” EMBO Reports 2025. Under Review.
Dallon, E.; Moran, H. M.; Chidambaran, S.; Kian,A.; Huang, B.; Fried, S.D.; DiRuggiero, J
“Investigation of the global translational response to oxidative stress in the model archaeon Haloferax volcanii reveals untranslated small RNAs with ribosome occupancy” mSphere 2025. Accepted.
Chakraborty, S.; Tripathi, S. J.; Vázquez-Rosa, E.; Wood, N.; Barker, S.; Chaubey, K.; Fujioka, H.; Miller, E.; Tyagi, R.; Vignane, T.; Sharma, S. M.; Thomas, B.; Weil, Z. M.; Nelson, R. J.; Orsburn, B. C.; Filipovic, M. R.; Fried, S. D.; Snyder, S. H.; Pieper, A. A.; Paul, B. D.
“Cystathionine γ-lyase Depletion Disrupts Accelerates Age-Dependent Neurodegeneration” Cell 2025. In Revision.
Xia, Y.; Amann, B.; Gillilan, R. E.; Sharma, P.; Sen, S.; Fleming, K. G.; Fried, S. D.*
“Phosphoglycerate Kinase Can Adopt a Topologically Misfolded Form that is More Stable than its Native State.” bioRxiv 2025. DOI: 10.1101/2025.06.24.661412. J. Am. Chem. Soc. 2025. Under Review.
Yadav, D.; Demiralp, I. I.; Fakler, M.; Fried, S. D.*
“Chaperone Dependency during Biogenesis Does Not Correlate with Chaperone Dependency during Refolding” bioRxiv 2025. DOI: 10.1101/2025.06.16.659923. Mol. Syst. Biol. 2025. In Revision.
Tarbox, H. E.; Branch, A.; Fried, S. D.
“Proteins with Cognition-Associated Structural Changes in a Rat Model of Aging Exhibit Reduced Refolding Capacity.” Science Advances 2025, 11, 28, adt3778.
2024
Manriquez-Sandoval, E.; Brewer, J.; Lule, G.; Lopez, S.; Fried, S. D.
“FLiPPR: A Processor for Limited Proteolysis (LiP) Mass Spectrometry Datasets Built on FragPipe” bioRxiv 2023. DOI: 10.1101/2023.12.04.569947v1 J. Proteome Res. 2024. Accepted.
To, P.; Bhagwat, A.; Tarbox, H. E.; Ecer, A.; Jamieson, Z.; Trcek, T.; Fried, S. D.
“Intrinsically Disordered Regions Promote Protein Refoldability and Facilitate Retrieval from Biomolecular Condensates. bioRxiv 2023. DOI: 10.1101/2023.06.25.546465 Nature 2024. Under Revision.
Dang, L. P.; Nissley, D. A.; Sitarik, I.; Vu, Q. V.; Jiang, Y.; To, P.; Xia, Y.; Fried, S. D.; Li, M. S.; O’Brien, E. P.
“Synonymous mutations can alter protein dimerization through localized interface misfolding involving self-entanglements” J. Mol Biol. 2024, 436,168487.
Baiz, C. R.; Berger, R. F.; Donald, K. J.; de Paula J. C.; Fried, S. D.; Rubenstein, B.; Stokes G. Y.; Takematsu, K.; Londercan G.
“Lowering Activation Barriers to Success in Physical Chemistry (LABSIP): A Community Project” J. Phys. Chem. A https://pubs.acs.org/doi/10.1021/acs.jpca.3c07015 2024, 128, 3-9.
Moran, H. M.; Manriquez-Sandoval, E.; Sharma, P.; Fried, S. D.; Gillilan, R. E.
“Proteome-Wide Assessment of Protein Structural Perturbations Under High Pressure” PRX Life 2024, 2, 033011.
2023
Xie, Q.; Vissamsetti, N.; Lee, S. O.; Fried, S. D.
“Secretion-catalyzed assembly of protein biomaterials on a bacterial membrane surface” Angew. Chemie. Int. Ed. https://doi.org/10.1002/anie.202305178 2023, 62, e202305178.
Jiang, Y.; Neti, S. S.; Pradhan, P.; Sitarik, I.; To, P.; Fried, S.D.; Booker, S.J.; O’Brien, E.P.
“How synonymous mutations alter enzyme structure and function over long time scales” https://www.nature.com/articles/s41557-022-01091-z Nature Chem. 2023, 15, 308–318
Faustino, A. M.; Sharma, P.; Manriquez-Sandoval, E.; Yadav, D.; Fried, S. D.
“Progress Toward Proteome-Wide Photo-Crosslinking to Enable Residue-Level Visualization of Protein Structures and Networks in vivo. DOI: /10.1021/acs.analchem.3c01369 Anal. Chem. 2023, 95, 10670–10685.
Makharov, M.; Sanchez Rocha, A. C.; Krystufek, R.; Dzmitruk, V.; Charnavets, T.; Faustino, A. M.; Lebl, M.; Fujishima, K.; Fried, S. D.; Hlouchova, K.
“Early selection of the amino acid alphabet was adaptively shaped by biophysical constraints of foldability” https://pubs.acs.org/doi/full/10.1021/jacs.2c12987 J. Am. Chem. Soc. 2023, 145, 5320–5329.
Vu, Q. V.; Sitarik, I.; Jiang, Y.; Yadav, D.; Shama, P.; Fried, S. D.; Li, M. S.; O’Brien, E. P.
“A Newly Identified Class of Protein Misfolding in All-atom Folding Simulations Consistent with Limited Proteolysis Mass Spectrometry bioRxiv 2022. DOI: 10.1101/2022.07.19.500586 v2. Nature Comm 2023. Under Review.
2022
Jacinto, M. P.; Fried, S. D.; Greenberg, M. M.,
“Intracellular Formation of a DNA Damage-Induced, Histone Post-Translational Modification Following Bleomycin Treatment” https://pubmed.ncbi.nlm.nih.gov/35467863/ J. Am. Chem. Soc. 2022, 144, 7600–7605.
Fried, S. D.; Booth, P. J.
“Editorial: Biophysics of co-translational protein folding,” https://www.frontiersin.org/articles/10.3389/fmolb.2022.1110432/full Front. Mol. Biosci. 2022, 9, 1110432.
Manriquez-Sandoval, E.; Fried, S. D.,
“DomainMapper: Accurate domain structure annotation including those with non-contiguous topologies” https://onlinelibrary.wiley.com/doi/full/10.1002/pro.4465, Protein Science 2022, 31, e4465.
Nissley, D. A.; Jiang, Y.; Trovato, F.; Sitarik, I. M.; Narayan, K.; To, P.; Fried, S. D.; O’Brien, E. P.
“Universal Protein misfolding intermediates can bypass the proteostasis network and remain soluble and non-functional.” https://www.nature.com/articles/s41467-022-30548-5 Nature Comm. 2022, 13, 3081.
To, P.; Xia, Y.; Lee, S. O.; Devlin, T.; Fleming, K. G.; Fried, S. D.
“A Proteome-Wide Map of Chaperone-Assisted Protein Refolding in a Cytosol-like Milieu” https://www.pnas.org/doi/full/10.1073/pnas.2210536119 Proc. Natl. Acad. Sci. USA 2022, 119, e2210536119.
Fried, S. D.; Fujishima, K.; Makarov, M.; Cherepashuk, I.; Hlouchova, K.
“Peptides Before and During the Nucleotide World: An Origins Story Emphasizing Cooperation between Proteins and Nucleic Acids. https://royalsocietypublishing.org/doi/10.1098/rsif.2021.0641 J. Roy. Soc. Interface. 2022, 19 (187), 20210641.
Nodelman, I. M.; Das, S.; Faustino, A. M.; Fried, S. D.; Bowman, G. D.; Armache, J.-P.
“Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.” https://www.nature.com/articles/s41594-021-00719-x Nature Struct. Mol. Biol.2022, 29, 121–129.
2021
Lee, S. O.; Fried, S. D.
“An Error Prone PCR Method for Short Amplicons.” https://pubmed.ncbi.nlm.nih.gov/34081928/ Anal. Biochem. 2021, 628, 114266.
To, P.; Whitehead, B.; Tarbox, H. E.; Fried, S. D.
“Non-Refoldability is Pervasive Across the E. coli Proteome.” https://pubs.acs.org/doi/10.1021/jacs.1c03270 J. Am. Chem. Soc. 2021, 143, 11435–11448.
Lee, S. O.; Xie, Q.; Fried, S. D.
“Optimized Loopable Translation as a Platform for the Synthesis of Repetitive Proteins.” https://pubs.acs.org/doi/10.1021/acscentsci.1c00574 ACS Central Science 2021, 7, 1736–1750.
2020
Marx, D. C.; Plummer, A. M.; Faustino, A. M.; Roskopf, M. A.; Leblanc, M. J.; Devlin, T.; Lessen, H. J.; Majumbdar, A.; Amann, B. T.; Flaming, P. J.; Kruegger, S.; Fried, S. D.; Fleming, K. G.
“SurA is a Cryptically Grooved Chaperone That Expands Unfolded Outer Membrane Proteins.” https://www.pnas.org/doi/full/10.1073/pnas.2008175117 Proc. Natl. Acad. Sci. USA. 2020, 117, 28026–28035.
Patents
Sharma, P.; Fried, S. D.; Faustino, A. M.
“Mass Spectrometry-Cleavable Photo-Crosslinkers for Proteome-Wide Photo-Crosslinking Mass Spectrometry” U.S. Provisional Patent Application No. 63/583,709
Lee, S. O.; Fried, S. D.
“A Loopable Translator for the Synthesis of Repetitive Proteins” U.S. Provisional Patent Application No. 63/374,949
Pre-Hopkins Pubs
Cervettini, D.; Tang, S.; Fried, S. D.; Willis, J. C. W.; Funke, L. F. H.; Chin, J. W.
“Rapid and Scalable Discovery and Evolution of Orthogonal Aminoacyl tRNA Synthetase/tRNA pairs.” Nature Biotech. 2020, 38, 989–999.
Wu, Y; Fried, S. D.; Boxer, S. G.
“A Preorganized Electric Field Leads to Minimal Reorientation in the Catalytic Reaction of Ketosteroid Isomerase.” J. Am. Chem. Soc. 2020, 142, 9993–9998.
Saggu, M.; Fried, S. D.; Boxer, S. G.
“Local and Global Electric Field Asymmetry in Photosynthetic Reaction Centers.” J. Phys. Chem. B 2019, 123, 1527–1536.
Schmied, W.; Tnimov, Z.; Uttamapinant, C.; Rae, C. D.; Fried, S. D.; Chin, J. W.
“Controlling Orthogonal Ribosome Subunit Interactions Enables Evolution of New Function.” Nature 2018, 564, 444-448.
Fried, S. D.; Boxer, S. G.
“Electric Fields and Enzyme Catalysis.” Annu. Rev. Biochem. 2017, 86, 387–415.
Wang, L.; Fried, S. D.; Markland, T. E.
“Proton Network Flexibility Enables Robustness and Large Electric Fields in the Ketosteroid Isomerase Active Site.” J. Phys. Chem. B 2017, 121, 9807–9815.
Boxer, S. G.; Fried, S. D.; Schneider, S. H.; Wu, Y.
“Electric Fields and Enzyme Catalysis.” Proceedings of the 24th International Solvay Conference on Chemistry, World Scientific Publishing Co. 2017.
Elliott, T. S.; Biano, A.; Townsley, F. M.; Fried, S. D.; Chin, J. W.
“Tagging and Enriching Proteins Enables Cell-Specific Proteomics.” Cell Chem. Biol. 2016, 23, 805–815.
Wu, Y.; Fried, S. D.; Boxer, S. G.
“Dissecting Proton Delocalization in an Enzyme’s Hydrogen Bond Network with Unnatural Amino Acids.” Biochemistry 2015, 54, 7110–7119.
Fried, S. D.; Schmied, W. H.; Uttamapinant, C.; Chin, J. W.
“Ribosome Subunit Stapling for Orthogonal Translation in E. coli.” Angew. Chem. Int. Ed. 2015, 54, 12791–12794.
Fried, S. D.; Boxer, S. G.
“Response to Comments on ‘Extreme electric fields power catalysis in the active site of ketosteroid isomerase.’” Science 2015, 349, 936.
Fried, S. D.; Boxer, S. G.
“Measuring Electric Fields and Noncovalent Interactions Using the Vibrational Stark Effect.” Accounts of Chemical Research 2015, 48, 998–1006.
Wang, L.; Fried, S. D.; Boxer, S. G.; Markland, T. E.
“Quantum Delocalization of Protons in the Hydrogen Bond Network of an Enzyme Active Site.” Proc. Natl. Acad. Sci. USA 2014, 111, 18454–18459.
Fried, S. D.; Bagchi, S.; Boxer, S. G.
“Extreme Electric Fields Drive Catalysis in the Active Site of Ketosteroid Isomerase.” Science 2014, 346, 1510–1514.
Mu, X.; Wang, Q.; Wang, L.-P.; Fried, S. D.; Piquemal, J.P.; Dalby, K. N.; Ren, P.
“Modeling Organochlorine Compounds and the σ-hole Effect Using a Polarizable Multipole Force Field.” J. Phys. Chem. B 2014, 118, 6456–6465.
Fried, S. D.; Wang, L.-P.; Boxer, S. G.; Ren, P.; Pande, V. S.
“Calculations of the Electric Fields in Liquid Solutions.” J. Phys. Chem. B 2013, 117, 16236–16248.
Fried, S. D.; Boxer, S. G.
“Thermodynamic Framework for Identifying Free Energy Inventories of Enzyme Catalytic Cycles.” Proc. Natl. Acad. Sci. USA 2013, 110, 12271–122276.
Fried, S. D.; Bagchi, S.; Boxer, S. G.
“Measuring Electrostatic Fields in Both Hydrogen Bonding and non-Hydrogen Bonding Environments Using Carbonyl Vibrational Probes.” J. Am. Chem. Soc. 2013, 135, 11181–11192.
Sigala, P. A.; Fafarman, A. T.; Schwans, J.P.; Fried, S. D.; Fenn, T. D.; Caaveiro, J. M. M.; Pybus, B.; Ringe, D.; Petsko, G. A.; Boxer, S. G.; Herschlag, D.
“Quantitative Dissection of Hydrogen Bond-Mediated Proton Transfer in the Ketosteroid Isomerase Active Site.” Proc. Natl. Acad. Sci. USA 2013, 110, E2552–E2561.
Bagchi, S.; Fried, S. D.; Boxer, S. G.
“A Solvatochromic Model Calibrates Nitriles’ Vibrational Frequencies to Electrostatic Field.” J. Am. Chem. Soc. 2012, 134, 10373–10376.
Levinson, N. M.; Fried, S. D.; Boxer, S. G.
“Solvent-induced Infrared Frequency Shifts in Aromatic Nitriles are Quantitatively Described by the Vibrational Stark Effect.” J. Phys. Chem. B 2012, 116, 10470–10476.
Fried, S. D.; Boxer, S. G.
“Evaluation of the Energetics of the Concerted Acid-Base Mechanism in Enzymatic Catalysis: The Case of Ketosteroid Isomerase.” J. Phys. Chem. B 2012, 116, 690–697.
Fried, S. D.; Kanan, M. W.; Nocera, D. G
“Bridged Bisindole Carboxylates as a Model for Oxidative O-O Homocoupling.” The Nucleus 2008, 86, 9, 14–20.
Rosenthal, J.; Chng, L. L.; Fried, S. D.; Nocera, D. G.
“Stereoelectronic Control of H2O2 Dismutation by Hangman Porphyrins.” Chem. Commun. 2007, 25, 2642–2644.